
The process of creating isoforms by processing the RNA differently is called “alternative splicing.” BLAST finds many different matches for the RNA sequence input that may be slightly different, or have gaps, due to alternative splicing.
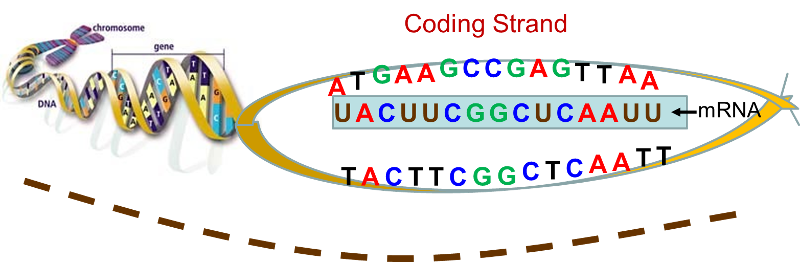
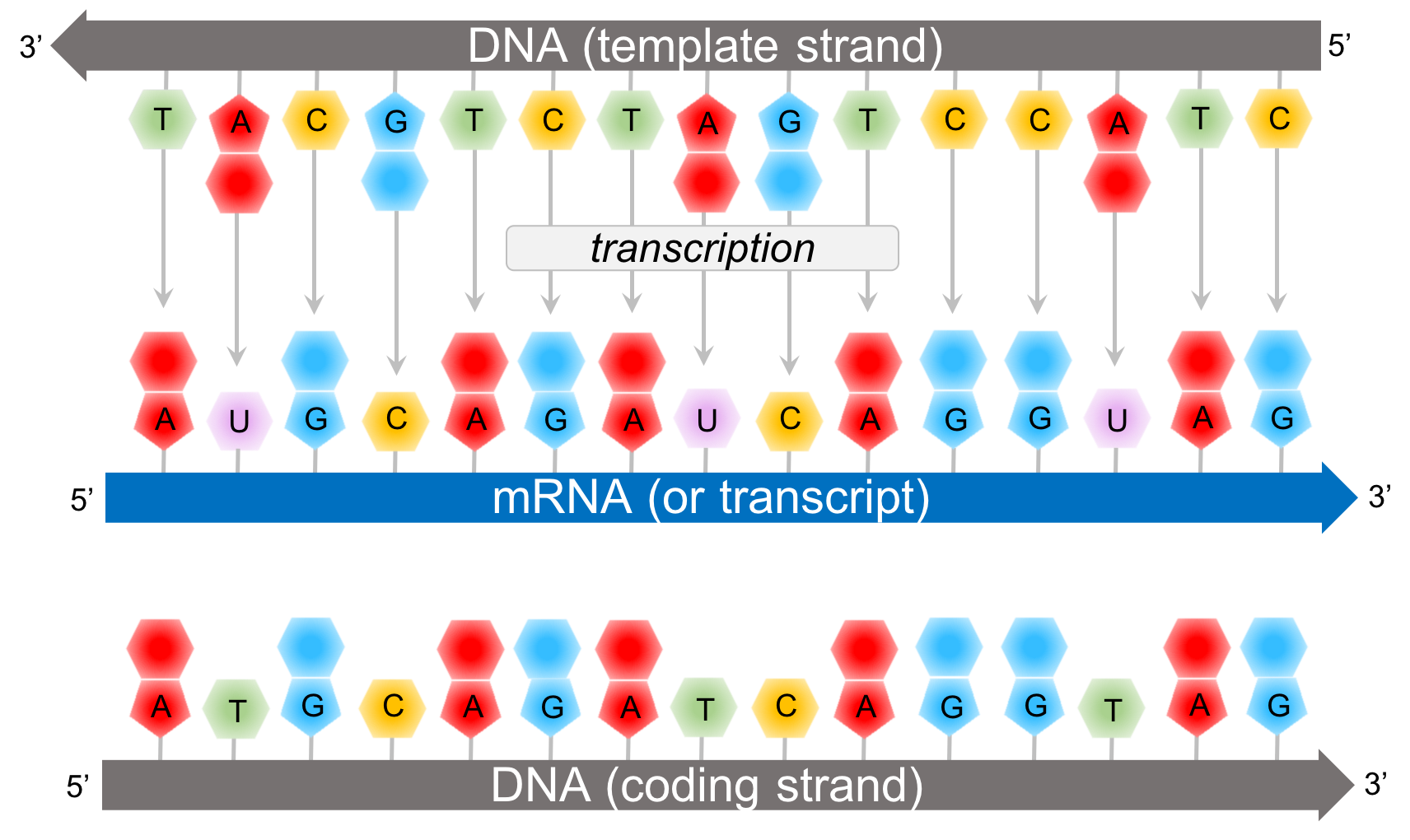
Depending on which introns are removed from the RNA, different “isoforms” (versions) can be created. So not all the RNA that is initially transcribed from the DNA makes it into the final RNA product. However, another element of complexity is that RNA is processed by having parts removed, i.e. the same as the negative strand, only with T’s replaced by U’s.īLAST finds all the matches it knows, from both positive and negative strand DNA and RNA this explains why some matches are Plus/Plus and others are Plus/Minus. For the example below, the reverse complementary RNA for the positive strand, read from the 5’ end, would be CAUCCU. The reverse complementary RNA for a positive strand DNA sequence will be identical to the corresponding negative strand DNA sequence. Question 2 adds the second step, reversing the sequence to give the proper 5'-3' orientation. Question 1 simulates the first step, finding the complementary sequence. The letters A, T, C and G represent the nucleotides or nitrogenous bases that make up a strand of DNA: A adenine. Give the DNA sequence that will pair with the following stretches of DNA. The RNA goes in the reverse direction compared to the DNA, but its base pairs still match (e.g. This usually involves reversing the sequence after writing it complementary to the one you are given. The 5’ end of the RNA will match up with the 3’ end of the DNA, and vice versa. When RNA is transcribed from the positive strand, it will be transcribed from the 3’ end towards the 5’ end of the DNA. One strand is designated the “positive” strand, and one is “negative.” DNA has a 5’ end and a 3’ end. During the meeting, the concept of reverse complementary DNA sequences was reviewed, in order to explain why BLAST will label some sequence matches as “Plus/Plus” and others as “Plus/Minus.” DNA has two strands (it is a double helix) that go in opposite directions. It also lists out information for each individual sequence match. DNA Sequence Reverse and Complement Online Tool With this tool you can reverse a DNA sequence, complement a DNA sequence or reverse and complement a DNA sequence Supports IUPAC ambiguous DNA characters. You may want to work with the reverse-complement of a sequence if it contains an ORF on the reverse strand. The entire IUPAC DNA alphabet is supported, and the case of each input sequence character is maintained. Lines from a FASTA file can be copy-pasted into BLAST, which will provide a visual summary of the matches that it has found for the input sequence. Reverse Complement converts a DNA sequence into its reverse, complement, or reverse-complement counterpart. FASTA files contain a name for each sequence and the order of the nucleotides in the sequence. The focus was on using Nucleotide BLAST to match RNA sequences from a file type called FASTA to RNA sequences in the NCBI database. During the March 24th meeting, members were introduced to BLAST, the Basic Local Alignment Search Tool ( ).


 0 kommentar(er)
0 kommentar(er)
